plot_guide_fidelity
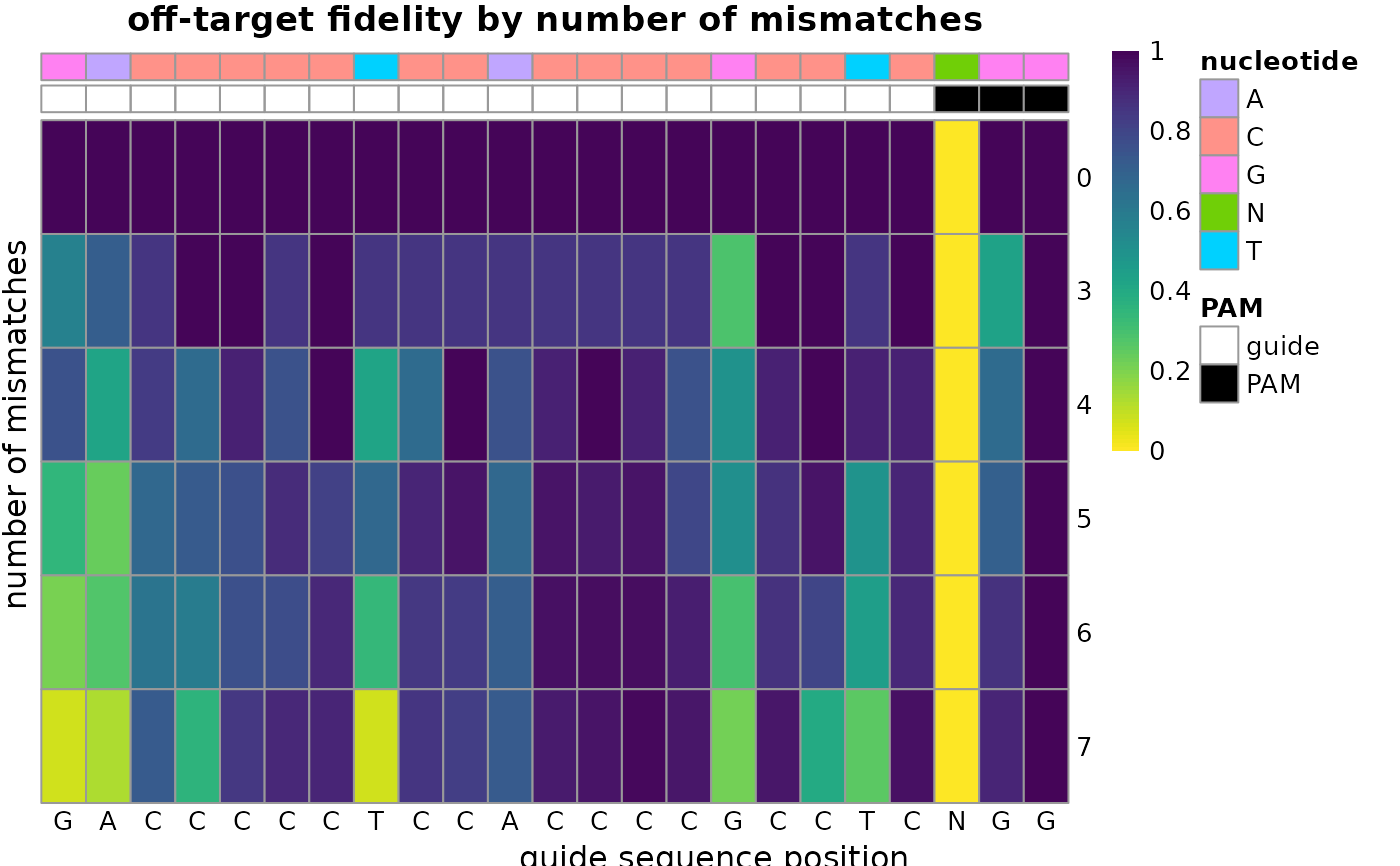
plot_guide_fidelity.RdShow, for each position of the guide, the fidelity of the offtarget sequence as the percentage of matching bases.
Details
Currently this function has the limitation that does only *fixed* matching, expecting the offtarget sequence and the guide to show the same character. See ?`Biostrings::lowlevel-matching` for more information.
Examples
if (FALSE) { # \dontrun{
offtargets <- breakinspectoR(
target =system.file("extdata/vegfa.chr6.bed.gz", package="breakinspectoR"),
nontarget=system.file("extdata/nontarget.chr6.bed.gz", package="breakinspectoR"),
guide ="GACCCCCTCCACCCCGCCTC",
PAM =c("NGG", "NAG"),
bsgenome ="BSgenome.Hsapiens.UCSC.hg38",
cutsiteFromPAM=3
)
} # }
data(breakinspectoR_examples)
plot_guide_fidelity(offtargets, guide="GACCCCCTCCACCCCGCCTC", pam="NGG")